6. Bessel Processes Part I#
The purpose of these notebooks (Bessel Processes Part I-III) is to provide an illustration of the Bessel Processes and some of their main properties.
In Part I, we introduce both Bessel and Squared Bessel processes with integer dimension \(d\geq 2,\) and study some of its main properties.
In Part II, we show that Bessel processes with integer dimension satisfy certain Stochastic Differential Equations (SDEs). This representation allows us to extend them to the non-integer case.
Finally, in Part III we illustrate both Bessel and Squared Bessel processes with general dimension \(\delta \geq 0\), and some of its main properties.
Before diving into the theory, let’s start by loading the libraries
matplotlib
together with the style sheet Quant-Pastel Light.
from aleatory.processes import BESProcess, BESQProcess
import matplotlib.pyplot as plt
quant_pastel = "https://raw.githubusercontent.com/quantgirluk/matplotlib-stylesheets/main/quant-pastel-light.mplstyle"
plt.style.use(quant_pastel)
plt.rcParams["figure.figsize"] = (12,6)
These tools will help us to make insightful visualisations of Bessel processes.
Important
In Part I, we will focus on the Bessel processes with integer dimension \(d\geq 2\), and starting at zero. In the following parts, we will cover how to generalise these two conditions.
6.1. Definition#
Let \(W =\{ W_t = (W^1_t, \cdots, W^d_t) : t\geq 0 \},\) be a \(d\)-dimensional standard Brownian Motion, for some integer \(d \geq 2\).
The Bessel process of dimension \(d\), is defined as the Euclidean norm of \(W\). That is
Similarly, the Squared Bessel process of dimension \(d\), is defined as the squared Euclidean norm of \(W\).
Notation
Hereafter, we will use the following notation:
\(BES_0^d\) : to denote a Bessel process of dimension \(d\) starting at zero, as defined in equation (6.1)
\(BES_0^d\) : to denote a Squared Bessel process of dimension \(d\) starting at zero, as defined in equation (6.2)
6.2. Simulation#
The previous definitons (in terms of a multidimensional Brownian Motion) allow us to simulate paths from Bessel processes of integer dimension easily as follows.
6.2.1. Bessel Process#
In order to simulate a path from a Bessel process with integer dimension \(d\geq 2\), let us start by taking a discrete partition over an interval \([0,T]\) for the simulation to take place. For simplicity, we are going to take an equidistant partition of size \(n\in \mathbb{N}\), over the interval \([0,T]\), i.e.:
Then, the goal is to simulate a path of the form
where \(W\) denotes a \(d\)-dimensional Brownian motion. To do this, we follow the next steps:
Step 1. For \(i=1, \cdots, d\):
Simulate a path of the form
\[\{ W_{t_j}^{i} , j=0,\cdots, n-1\},\]That is, we simulate from \(d\) independent standard Brownian motions. See Brownian Motion for details on how to simulate from a standard Brownian motion. This allows us to construct the vectors
Step 2. For \(j=1,\cdots\ n\):
Take the norm of such vectors, that is
\[\begin{equation*} X_{t_j} = \sqrt{ \sum_{i=1}^d (W_{t_j}^{i})^2 }. \end{equation*}\]
# Snippet to simulate a path from a Bessel process with integer dimension
from aleatory.processes import BrownianMotion
import numpy as np
d = 3
T = 1.0
n = 100
times = np.linspace(0, T, n) # Partition of the interval [0,T]
brownian = BrownianMotion(T) # A Brownian Motion instance
brownian_samples = [brownian.sample_at(times) for _ in range(d)] # Step 1. Building the vectors (W_{t_j}^1 , ... W_{t_j}^d)
bessel_path = np.array([np.linalg.norm(coord) for coord in zip(*brownian_samples)]) # Step 2. Taking the Euclidian norm
Now, let’s plot our simulated path!
plt.plot(times, bessel_path, 'o-', lw=1)
plt.title('Bessel Process Path')
plt.show()
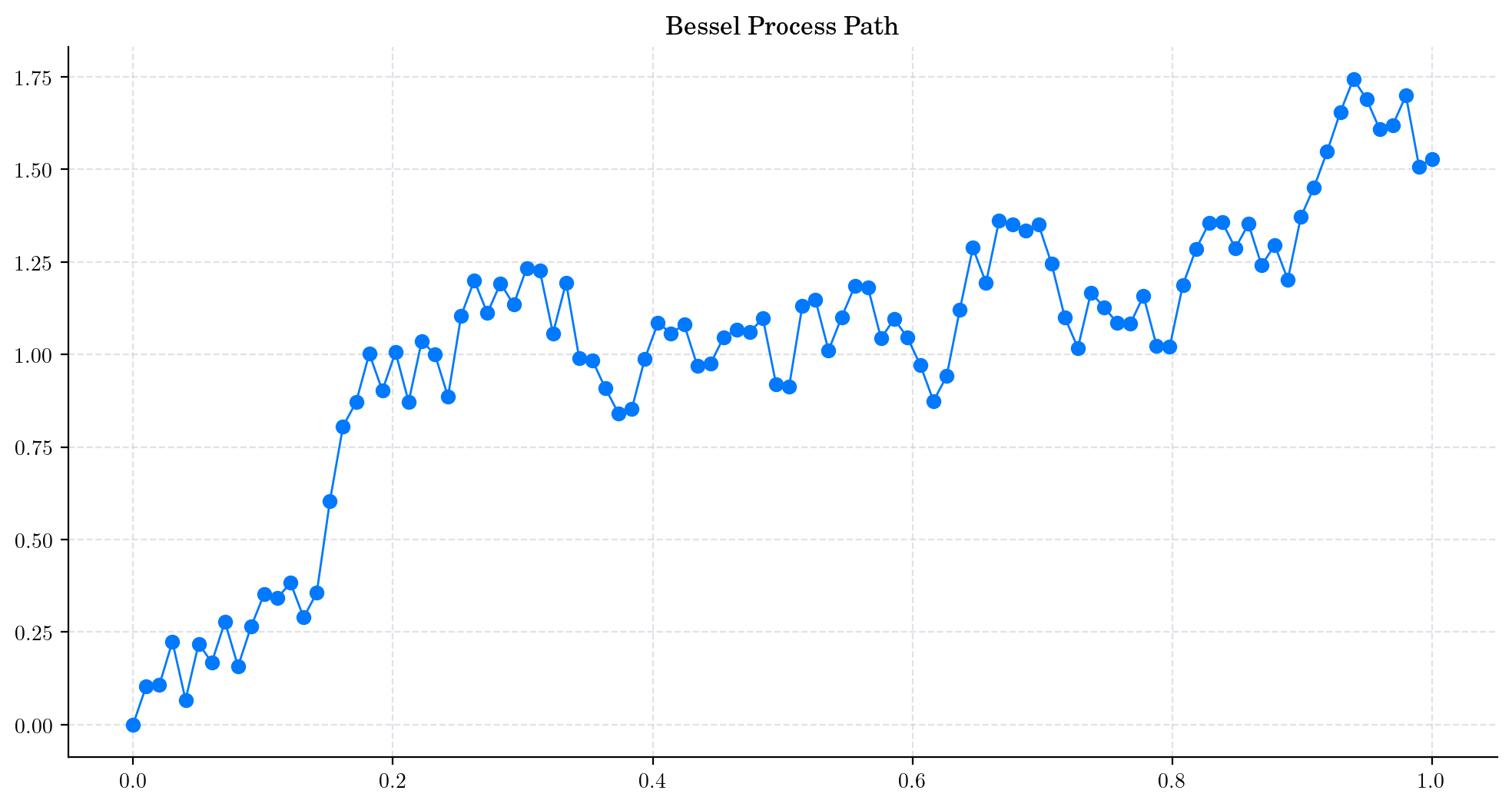
Note
In this plot, we are using a linear interpolation to draw the lines between the simulated points.
6.2.2. Squared Bessel Process#
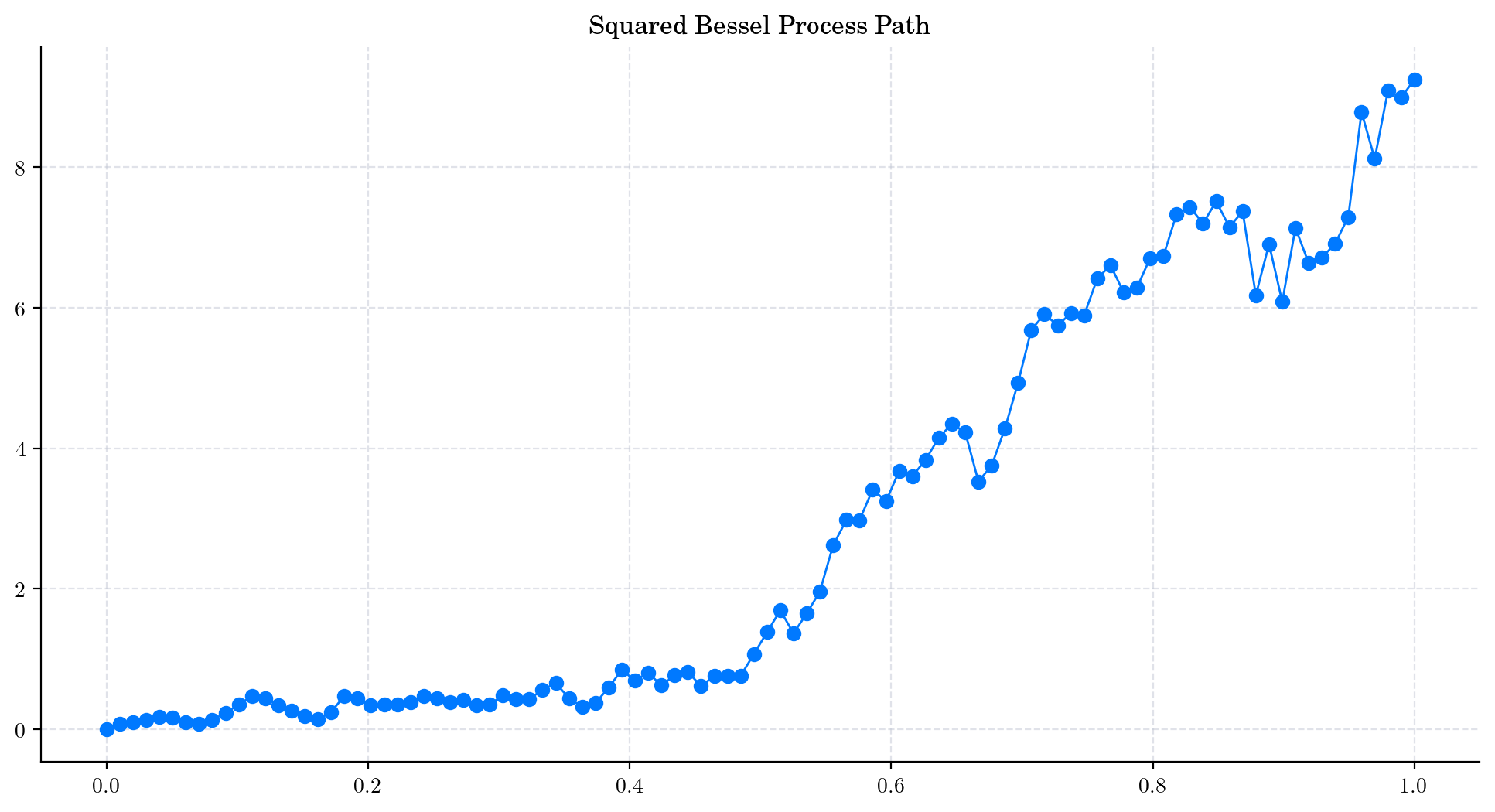
We can follow the same logic to simulate a path from a Squared Bessel process. The only differences is the calculation in Step 2. Instead of calculating the norm we just get the squared norm, i.e.:
Let’s write this in Python.
# Snippet to simulate a path from a Squared Bessel process with integer dimension
d = 4
T = 1.0
n = 100
times = np.linspace(0, T, n) # Partition of the interval [0,T]
brownian = BrownianMotion(T) # A Brownian Motion instance
brownian_samples = [brownian.sample_at(times) for _ in range(d)] # Step 1. Building the vectors (W_{t_j}^1 , ... W_{t_j}^d)
bessel_path = np.array([(np.linalg.norm(coord))**2 for coord in zip(*brownian_samples)]) # Step 2 (Modified). Taking the Square of the Euclidian norm
plt.plot(times, bessel_path, 'o-', lw=1) # Plot the path
plt.title('Squared Bessel Process Path')
plt.show()
6.2.3. Simulating and Visualising Paths#
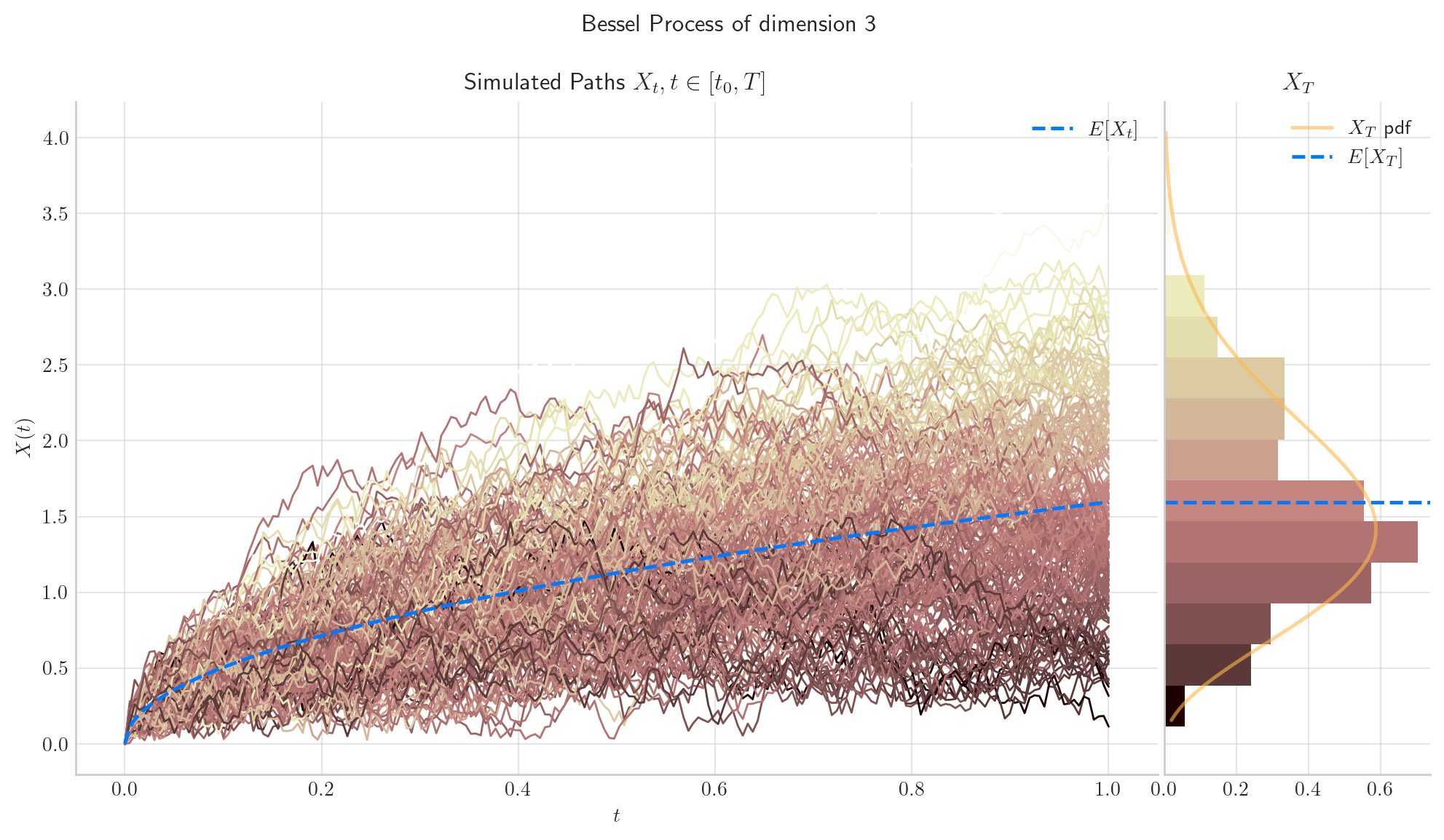
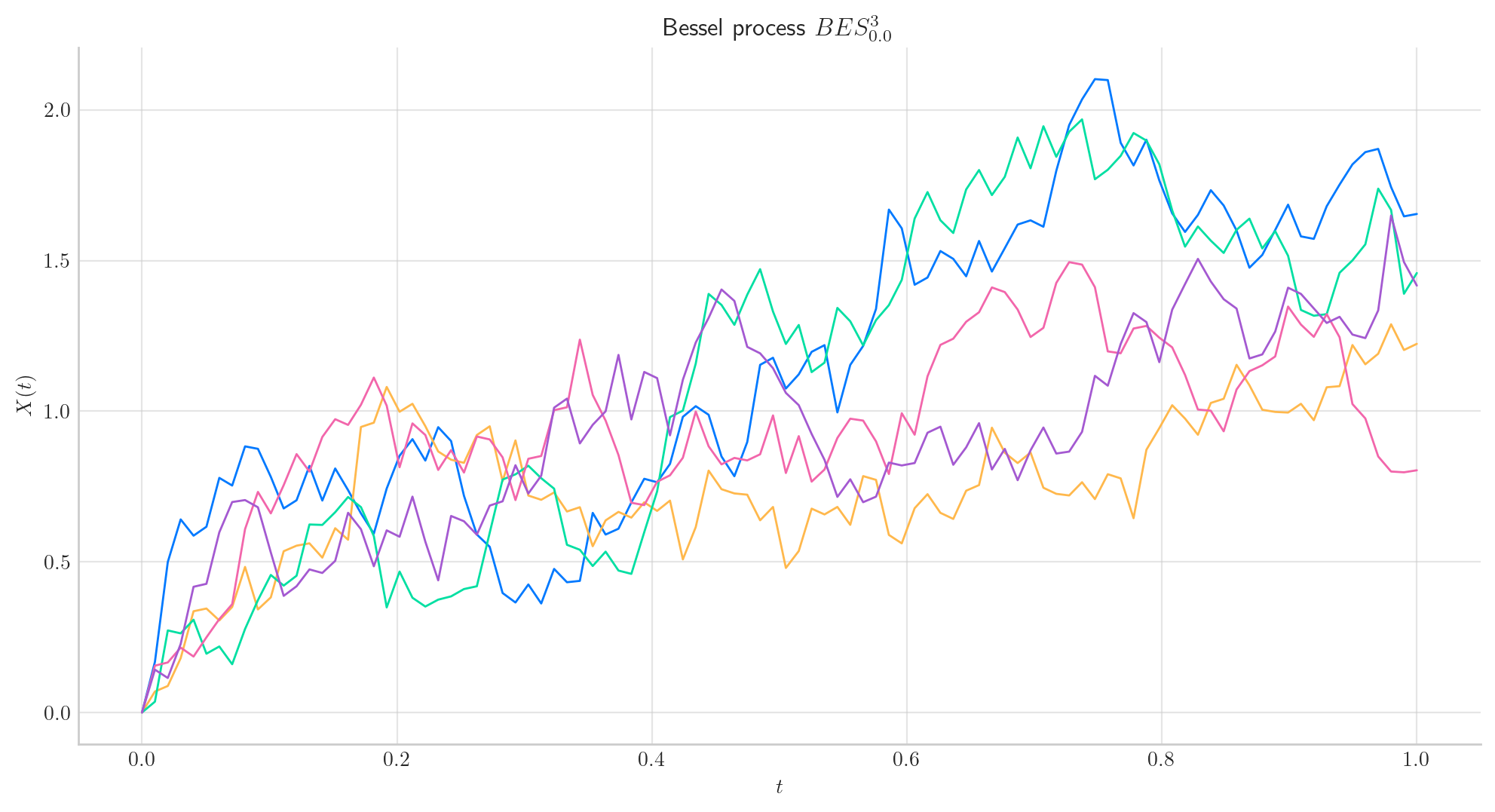
To simulate several paths from Bessel and Squared Bessel processes and visualise them we can use the methods simulate and plot from the aleatory library.
Let’s simulate 5 paths from a Bessel Process of dimension 3, over the interval \([0,1]\) using a partition of 100 points.
Tip
Remember that the number of points in the partition is defined by the parameter \(n\), while the number of paths is determined by \(N\).
The following snippet simulates the paths and returns them as a numpy array object.
# Snippet to Simulate N paths from the Bessel process
from aleatory.processes import BESProcess
bes = BESProcess(dim= 3)
paths = bes.simulate(n=100, N=5)
If we want to simulate and visualise the paths, we will use the method plot from the aleatory library.
# Snippet to Simulate and Visualise the paths from the Bessel process
from aleatory.processes import BESProcess
bes = BESProcess(dim= 3)
fig = bes.plot(n=100, N=5)
Similarly, let’s simulate 5 paths from a Squared Bessel process of dimension 6, over the interval \([0,1]\) using a partition of 100 points.
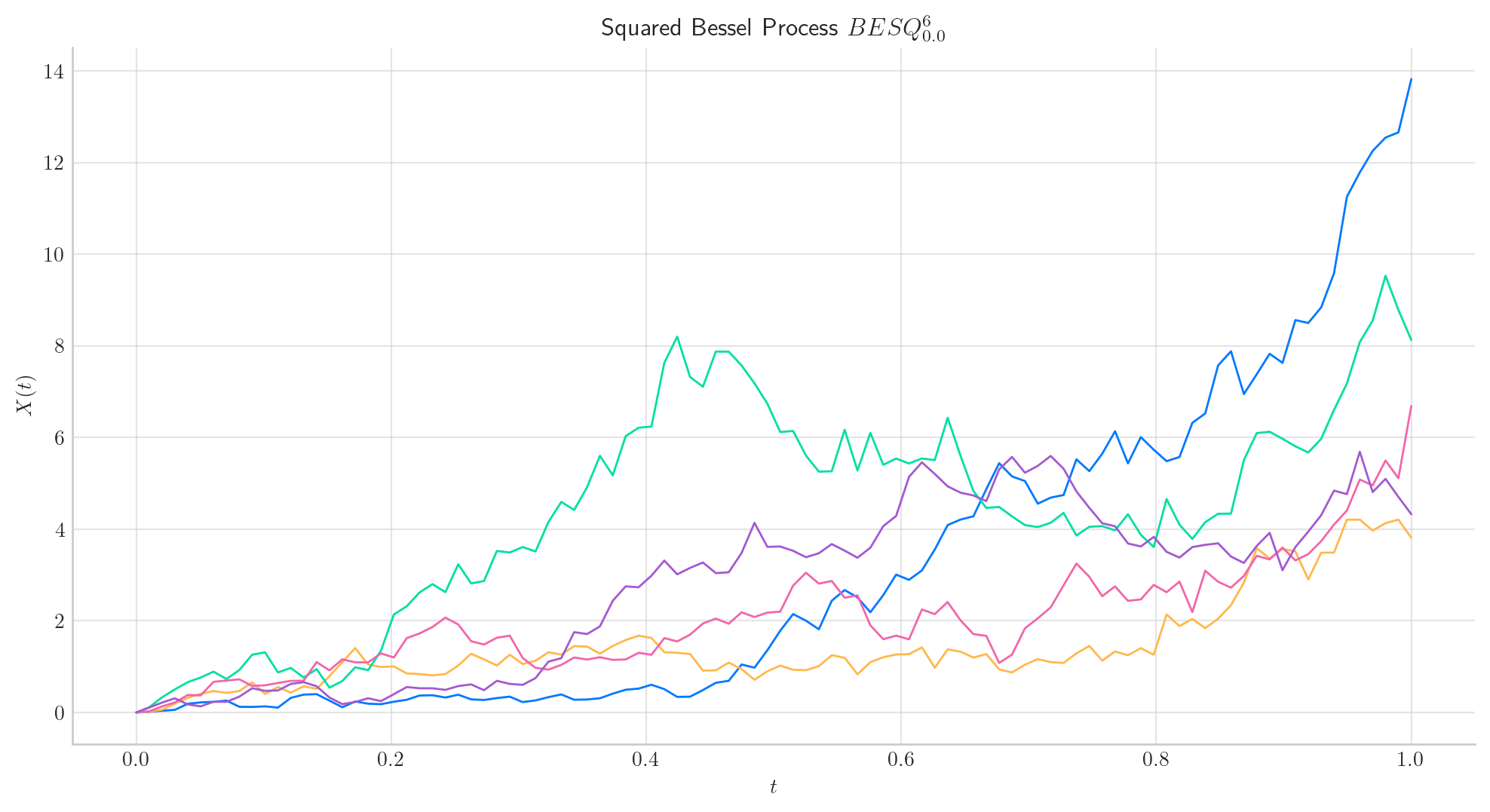
Note
In all plots we are using a linear interpolation to draw the lines between the simulated points.
6.3. Marginal Distributions#
For any given \(t\geq 0\), the definition of a \(d\)-dimensional Brownian Motion implies that each component \(W^i_t\), follows a normal distribution \(\mathcal{N}(0,t)\). Moreover, these components are independent of each other.
Hence, we have
i.e.; the marginal distribution of a Bessel process \(BES_0^d\) (with integer dimension \(d\geq2\)) is a scaled Chi random-variable with \(d\)-degrees of freedom.
Similarly,
that is, the marginal distribution of a Squared Bessel process \(BESQ_0^2\) (with integer dimension \(d\geq 2\)) is a scaled Chi-squared with \(d\)-degrees of freedom.
Note
The number of degrees of freedom \(d\) does not depend on \(t\), which means that the number of degrees of freedom remains constant as the process evolves.
6.3.1. Expectation and Variance#
6.3.1.1. Bessel#
For each \(t>0\), the marginal distribution \(X_t|X_0=0\) from a Bessel process \(BES_0^d\), with integer dimension \(d\geq 2\), satisfies
and
6.3.1.1.1. Python Implementation#
For given \(d\geq 2, t\geq 0\), we can implement the above formulas for the expectation, and variance, as follows.
from scipy.special import gamma
from math import sqrt
d = 3
t= 2
expectation = sqrt(2*t)*gamma((d+1)/2)/gamma(d/2)
variance = t*d - expectation**2
print(f'For d={d}' , f't={t}', sep=", ")
print(f'E[X_t] = {expectation: .6f}')
print(f'Var[X_t] = {variance :.14f}')
For d=3, t=2
E[X_t] = 2.256758
Var[X_t] = 0.90704182105935
Alternatively, we can use the method get_marginal to obtain the marginal distribution and the get its mean and variance
from aleatory.processes import BESProcess
d = 3
t= 2
bes = BESProcess(dim=d)
marginal = bes.get_marginal(t=t)
print(marginal.mean())
print(marginal.var())
2.2567583341910256
0.9070418210593473
6.3.1.2. Squared Bessel#
For each \(t>0\), the conditional marginal \(Y_t|Y_0=0\) from a Squared Bessel process starting from zero with integer dimension \(d\geq 2\), satisfies
and
6.3.1.2.1. Python Implementation#
For given \(d\geq 2, t\geq 0\), we can implement the above formulas for the expectation, and variance, as follows.
d = 3
t= 2
expectation = t*d
variance = (t**2)*(2*d)
print(f'For d={d}' , f't={t}', sep=", ")
print(f'E[Y_t] = {expectation: .6f}')
print(f'Var[Y_t] = {variance :.6f}')
# from aleatory.processes import BESProcess
bes = BESQProcess(dim=d)
marginal = bes.get_marginal(t=t)
print(marginal.mean())
print(marginal.var())
For d=3, t=2
E[Y_t] = 6.000000
Var[Y_t] = 24.000000
6.0
24.0
6.3.2. Probability Density Functions#
The probability density function (pdf)of the marginal distribution \(X_t\), from a Bessel process with integer dimension \(d\geq 0\), is given by the following expression
The probability density function (pdf)of the marginal distribution \(Y_t\), from a Squared Bessel process with integer dimension \(d\geq 0\), is given by the following expression
6.3.2.1. Visualisation#
To visualise these probability density functions in Python, we can use the method get_marginal from the aleatory library. Let’s consider a Bessel process BESProcess(dim=3) and plot the density function of the marginal \(X_1\)
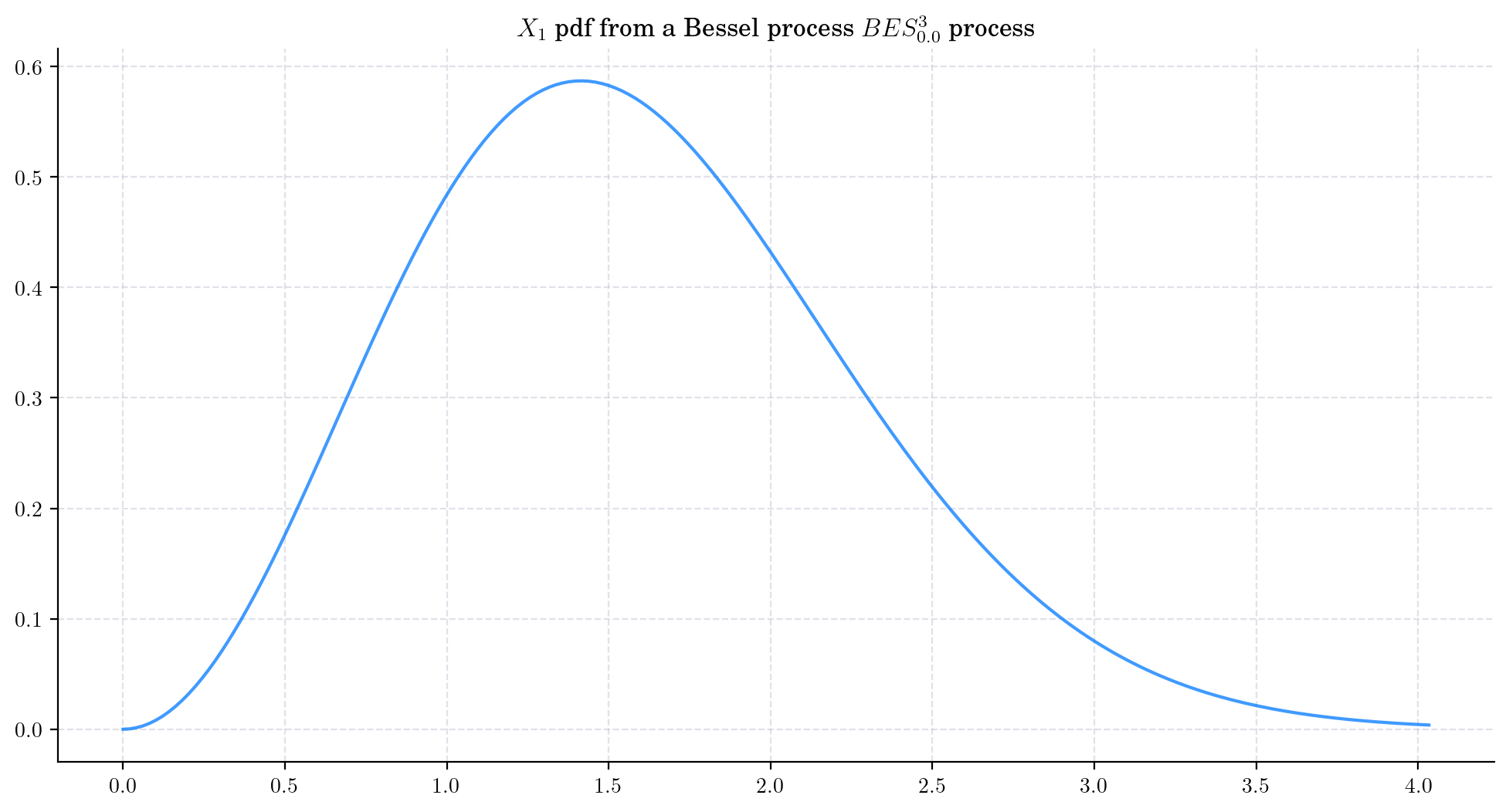
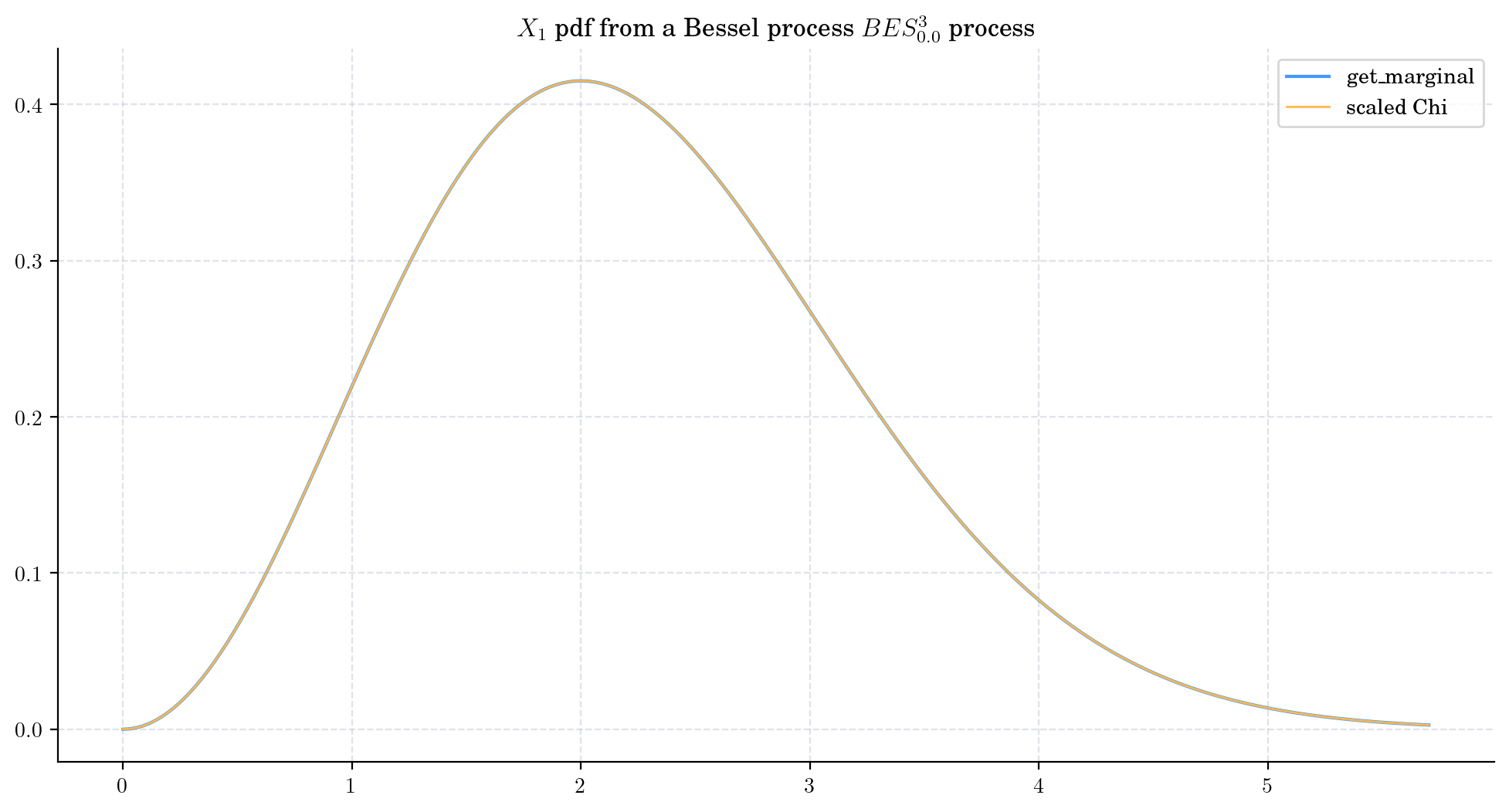
We can also use do this by using the fact that \(X_1\) follows a Chi distribution with parameter 3. You can try this alternative method by running the following code.
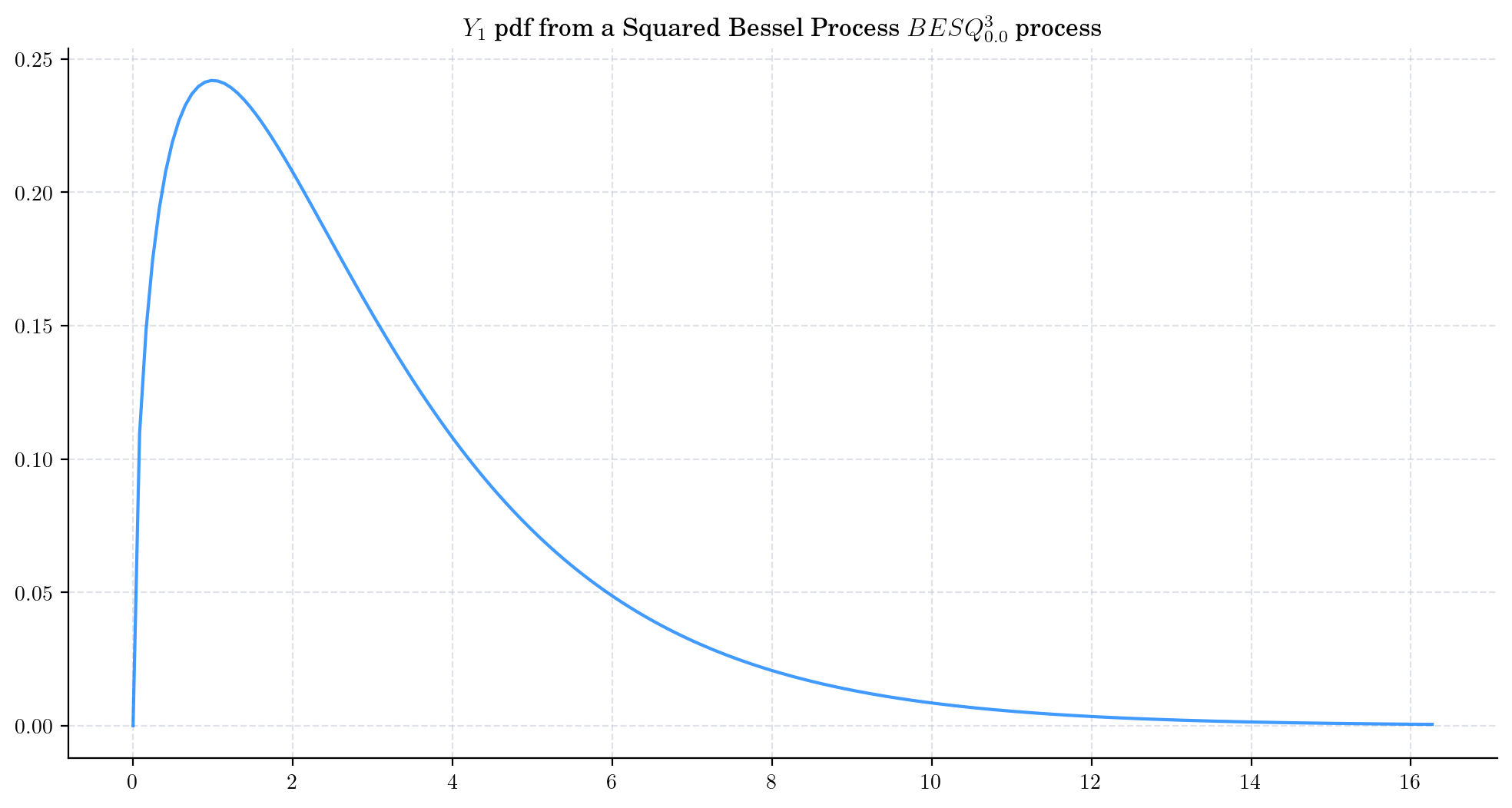
Similarly, we can take a Squared Bessel process BESQProcess(dim=3) and plot the density function of \(Y_1\).
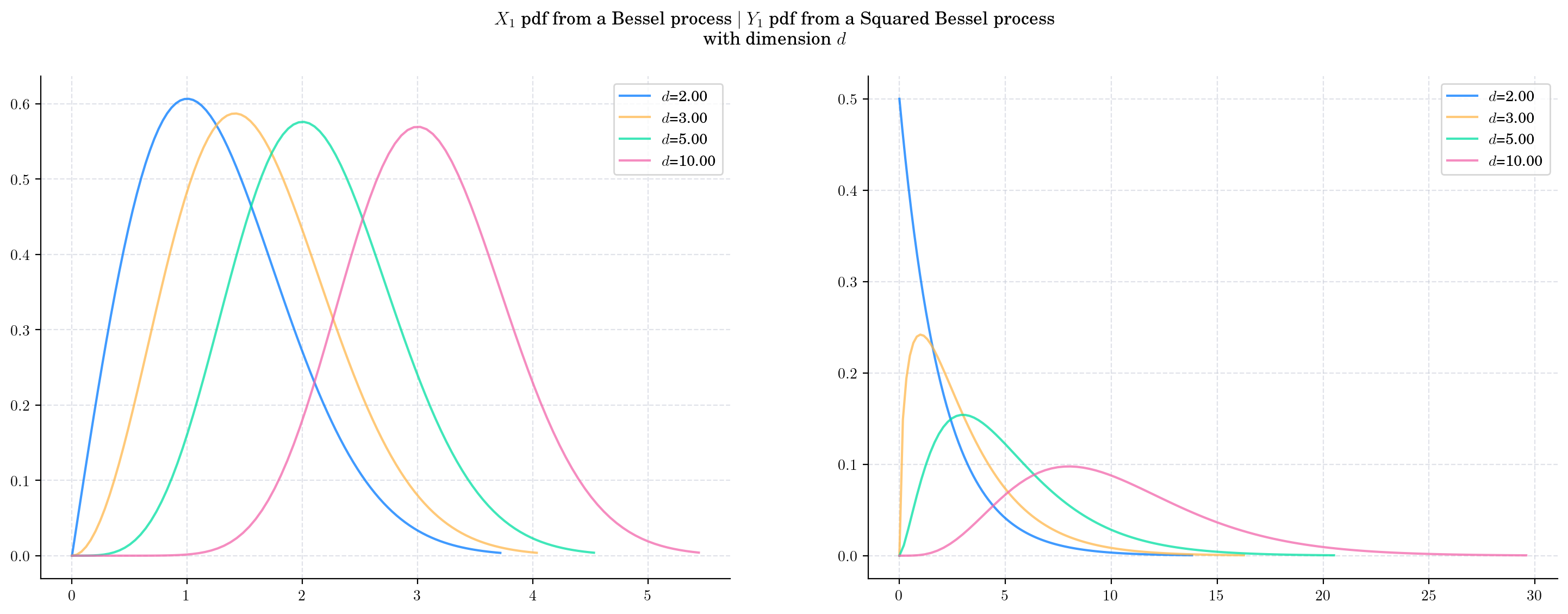
Next, we vary the value of the dimension \(d\) and plot the corresponding pdfs. As \(d\) increases:
The density from the Bessel marginal moves towards the right and becomes more symmetric.
The density from the Squared Bessel marginal becomes wider and the shape changes as well.
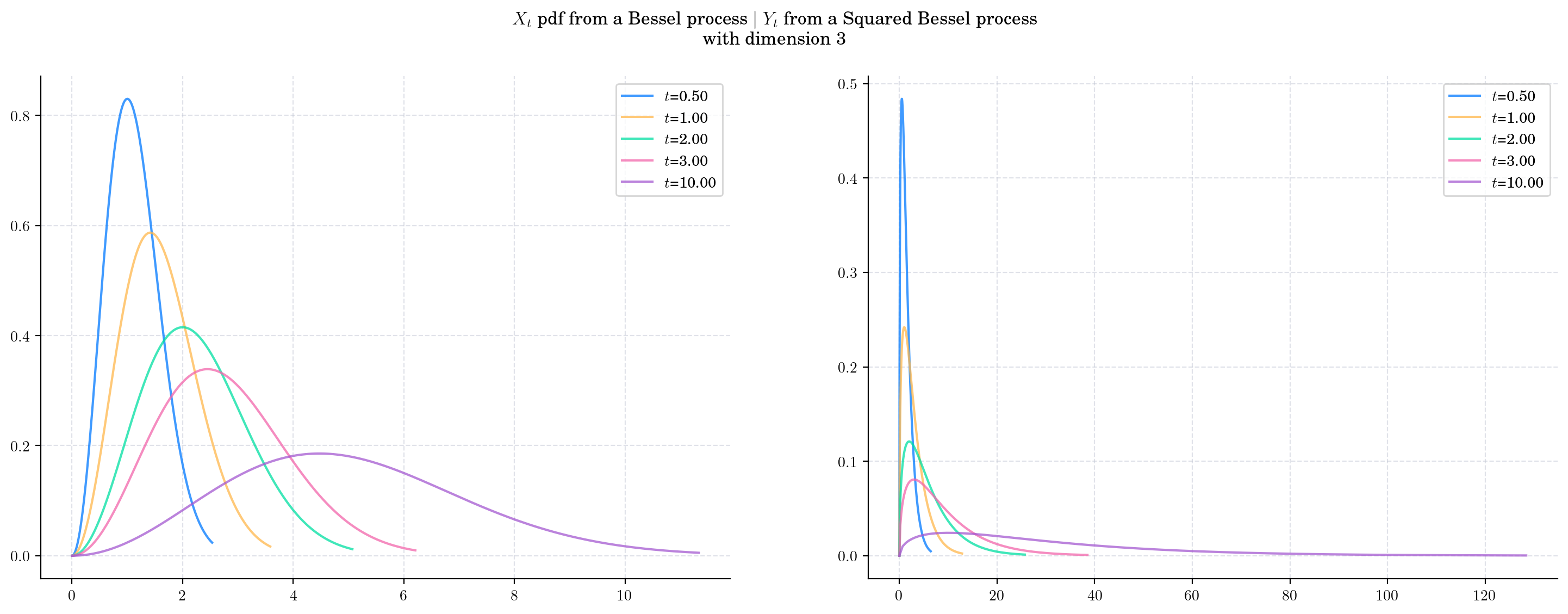
Finally, we fix the dimension (to 3) and vary the value of time \(t\) and plot the corresponding pdfs. As \(t\) increases:
The density from the Bessel marginal moves towards the right and becomes wider. This reflects the fact that both the mean and the variance are increasing.
The density from the Squared Bessel marginal becomes wider very rapidly as \(t\) grows.
6.4. Long Time Behaviour#
6.4.1. Expectation and Variance#
6.4.1.1. Bessel#
The conditional marginal from a Bessel process \(BES_0^d\), with integer dimension \(d\geq 2\), satisfies
and
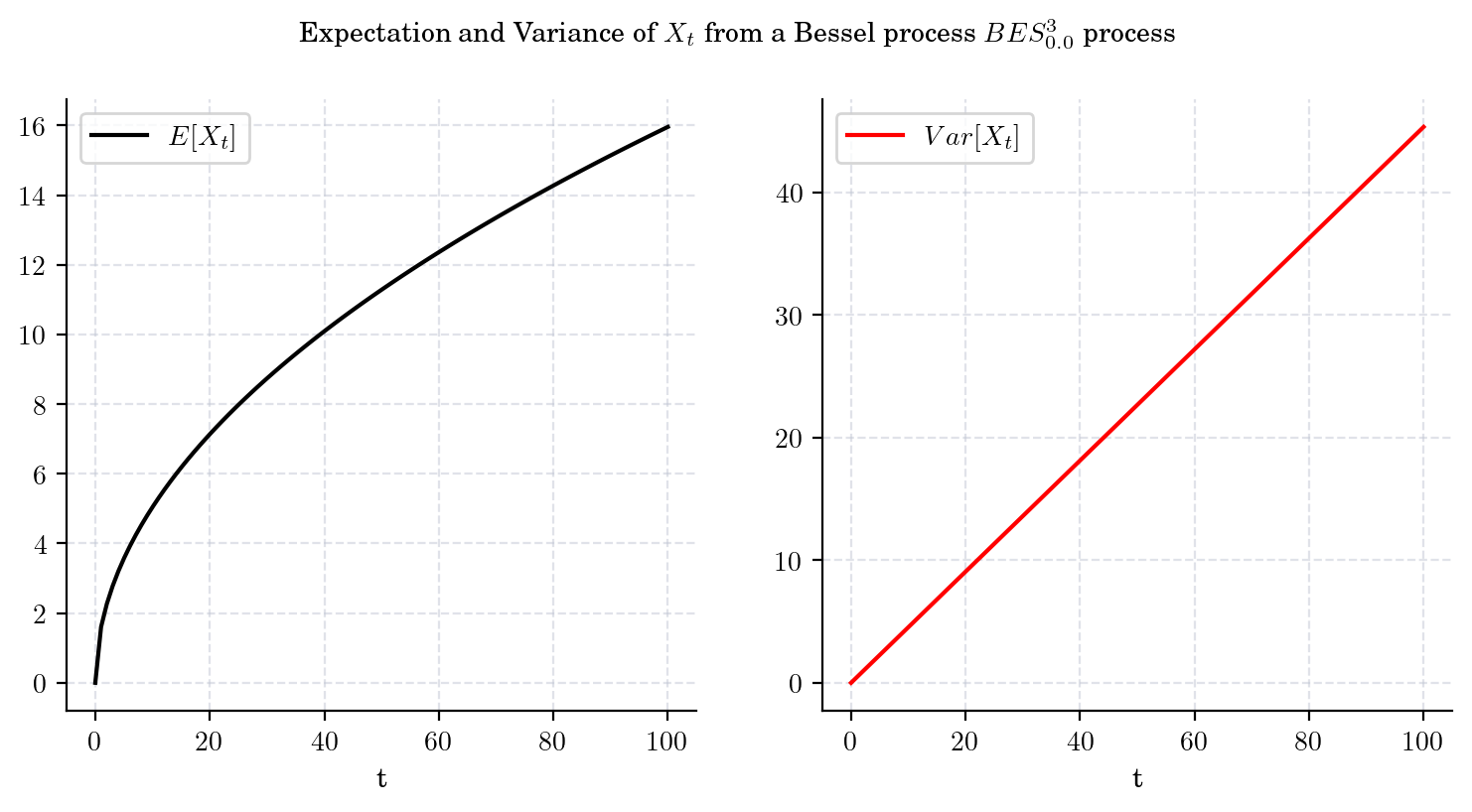
That is, both the mean and the variance tend to infinity as \(t\) grows. Besides, these expressions imply that the variance grows faster (at linear rate with respect to \(t\)) than the mean. The next plot illustrates these observations.
draw_mean_variance(dim=3, T=100)
6.4.1.2. Squared Bessel#
The conditional marginal from a Squared Bessel process \(BESQ_0^d\), with integer dimension \(d\geq 2\), satisfies
and
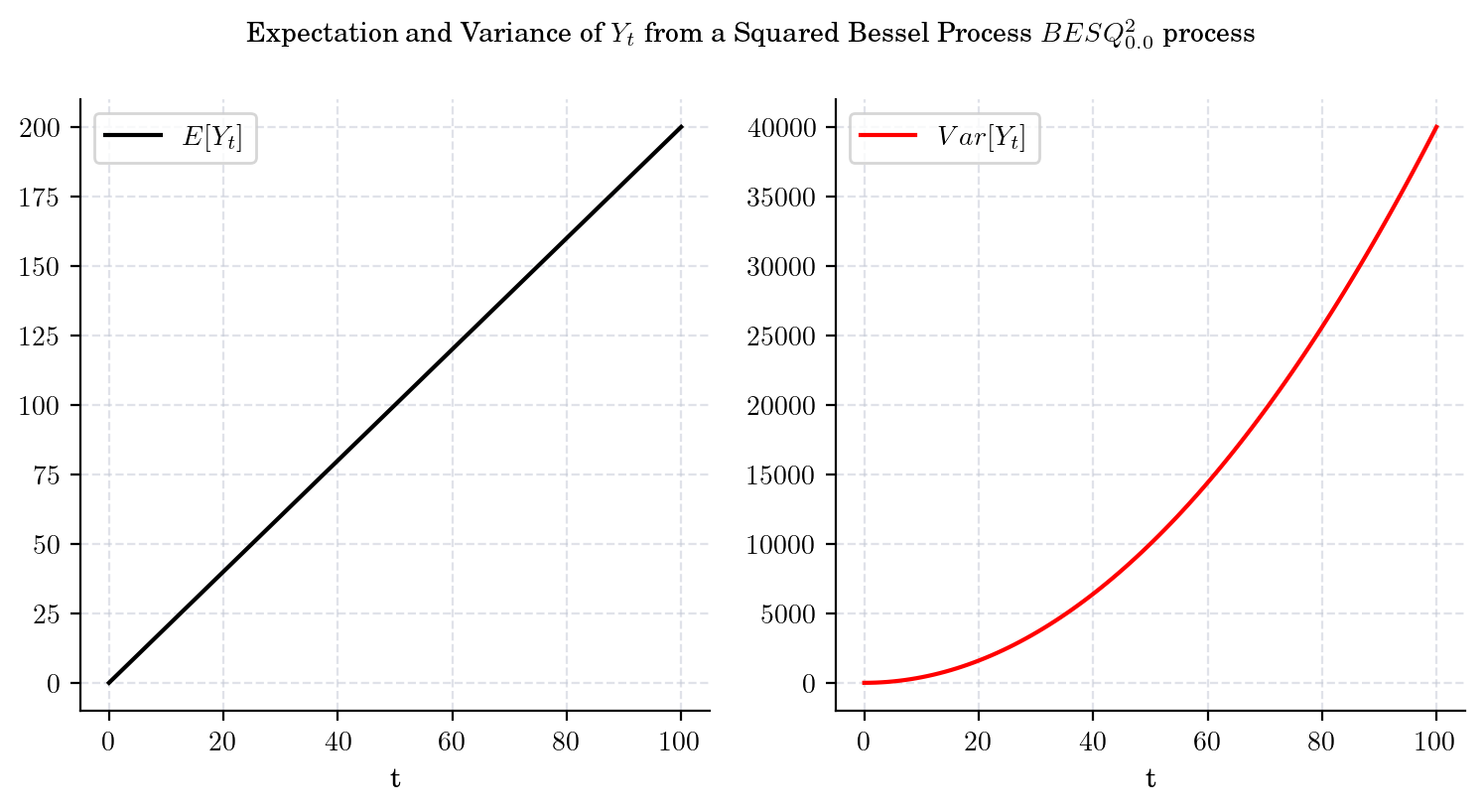
That is, both the mean and the variance tend to infinity as \(t\) grows. Besides, from these equations we can conclude that the variance grows faster than the mean. The next plot illustrates these observations.
draw_mean_variance(dim=2, T=100)
6.4.2. Marginal#
From equations (6.3) and (6.4) we have:
and
The following charts illustrate this behaviour.
In particular, we can see that both the probability density functions corresponding to \(X_t\), and \(Y_t\), become flatter and wider as \(t\) grows.
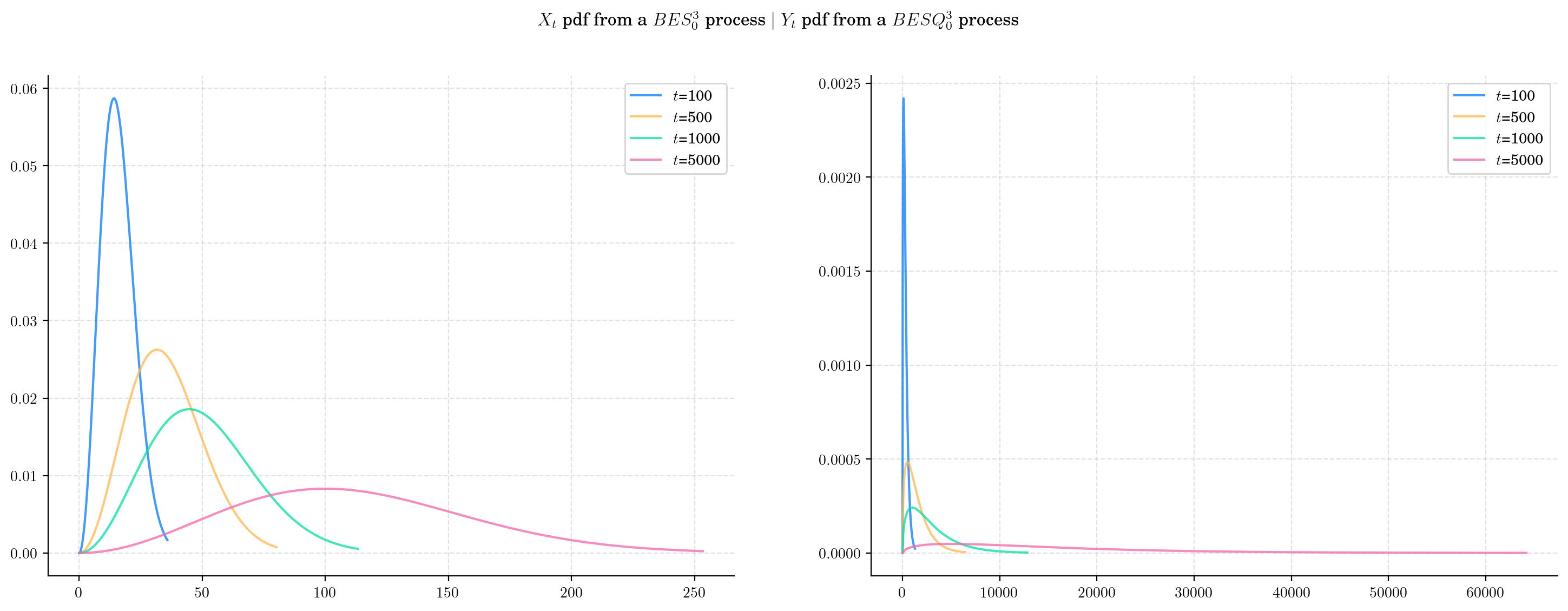
The following two charts show simulations from processes \(BES_0^3,\) and \(BESQ_0^3\), over the interval \([0,5000]\). In both cases, the plots illustrate the described long time behaviour.
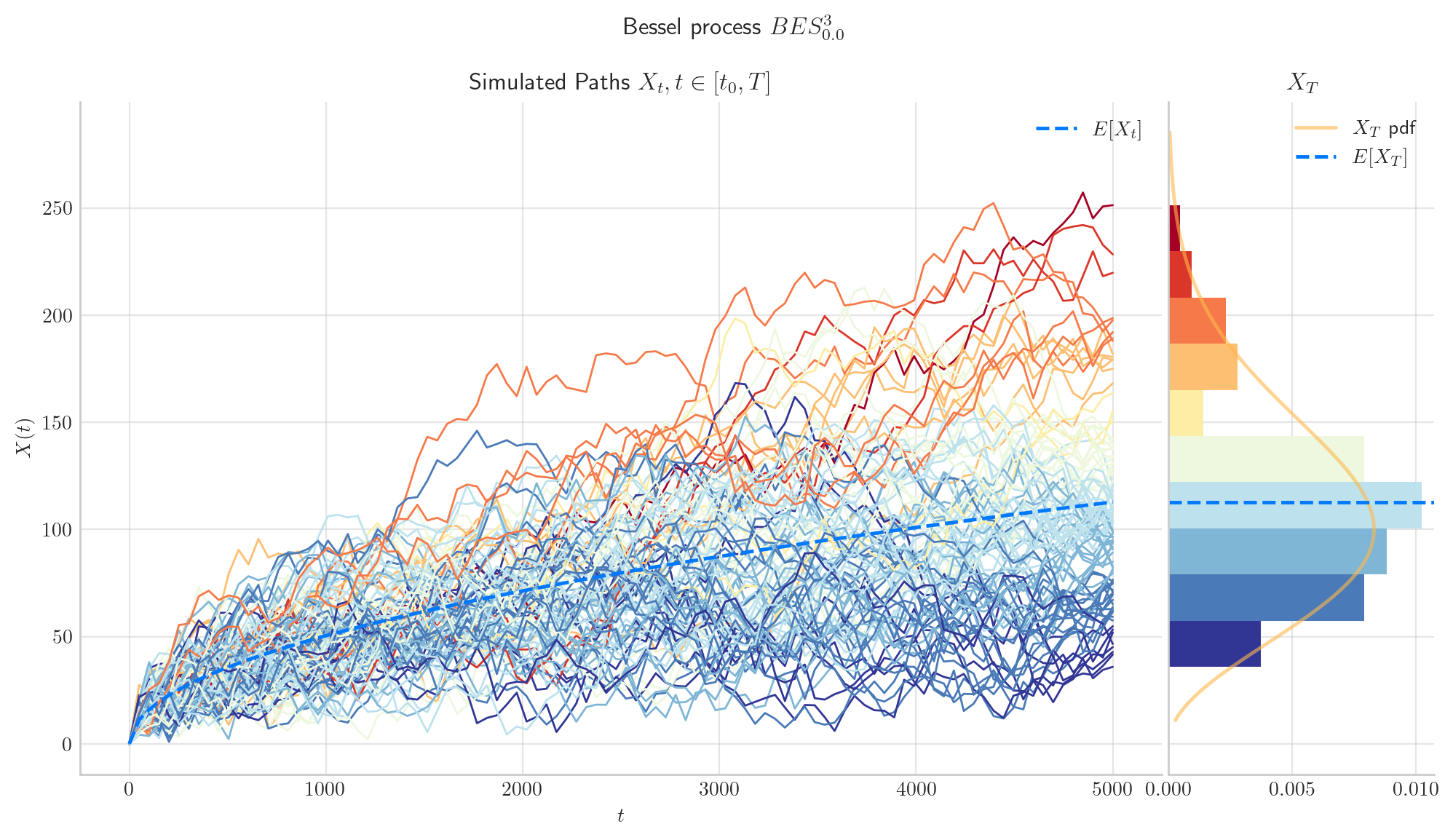
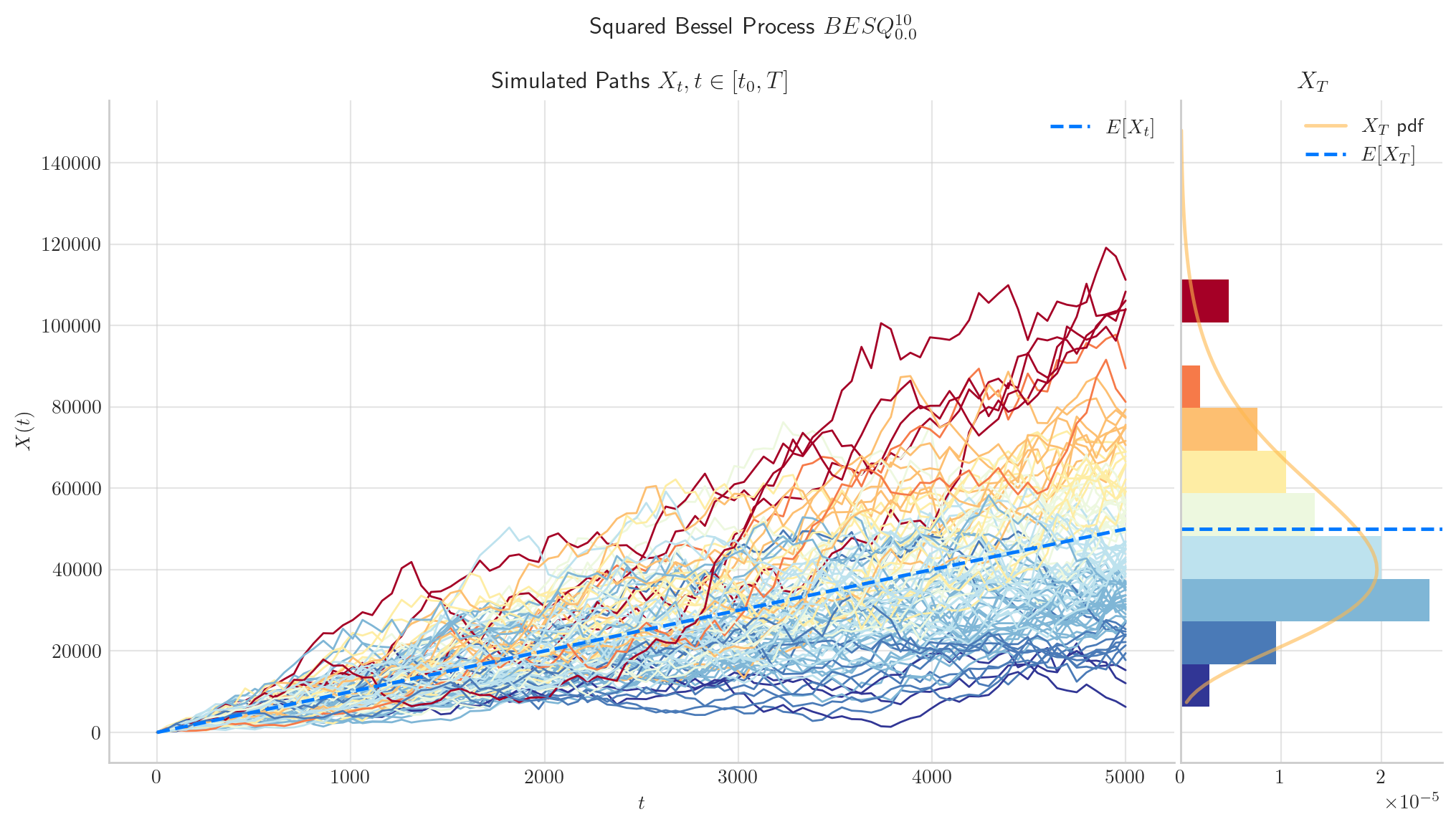
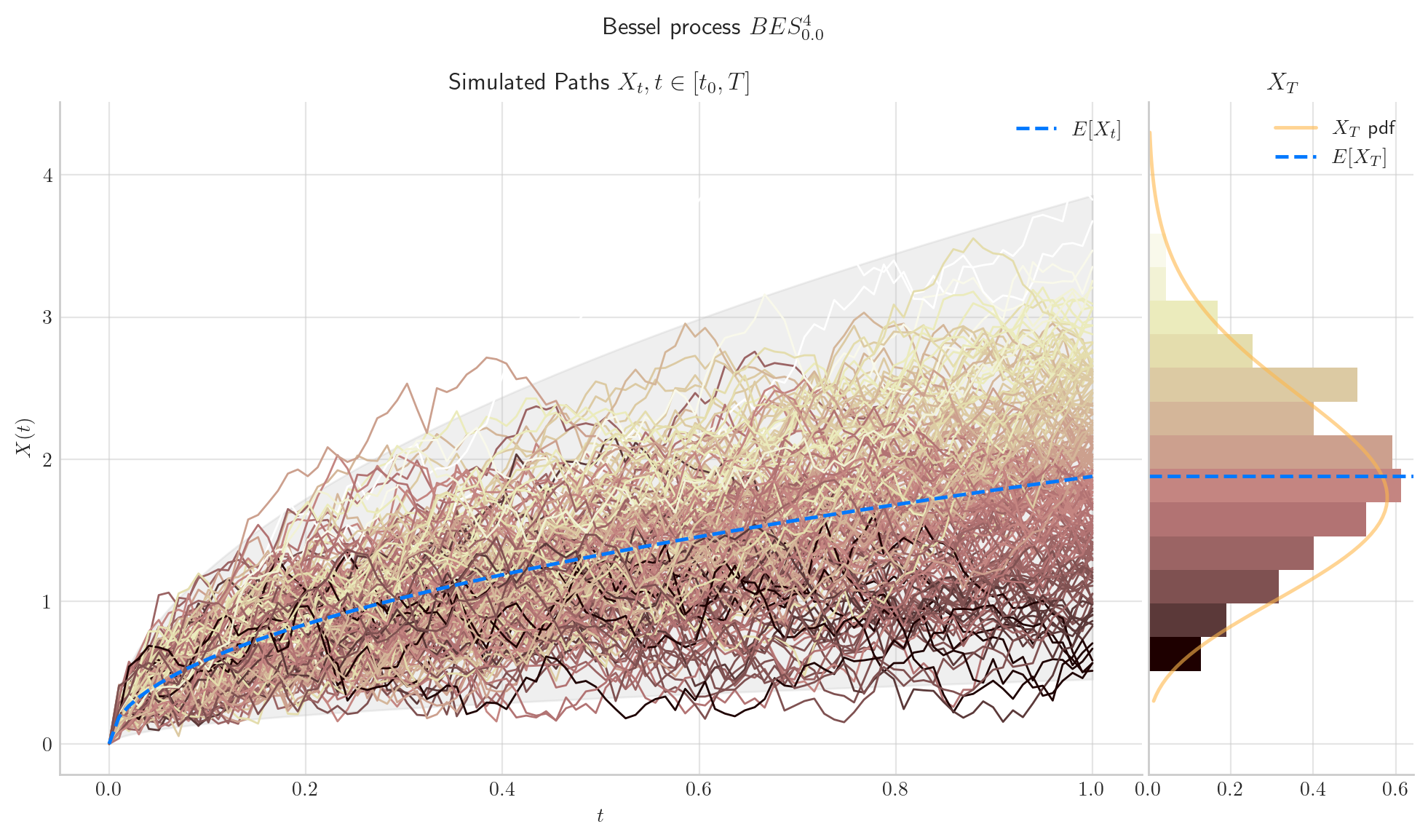
6.5. Final Visualisations#
To finish this note, let us take a look at some simulations.
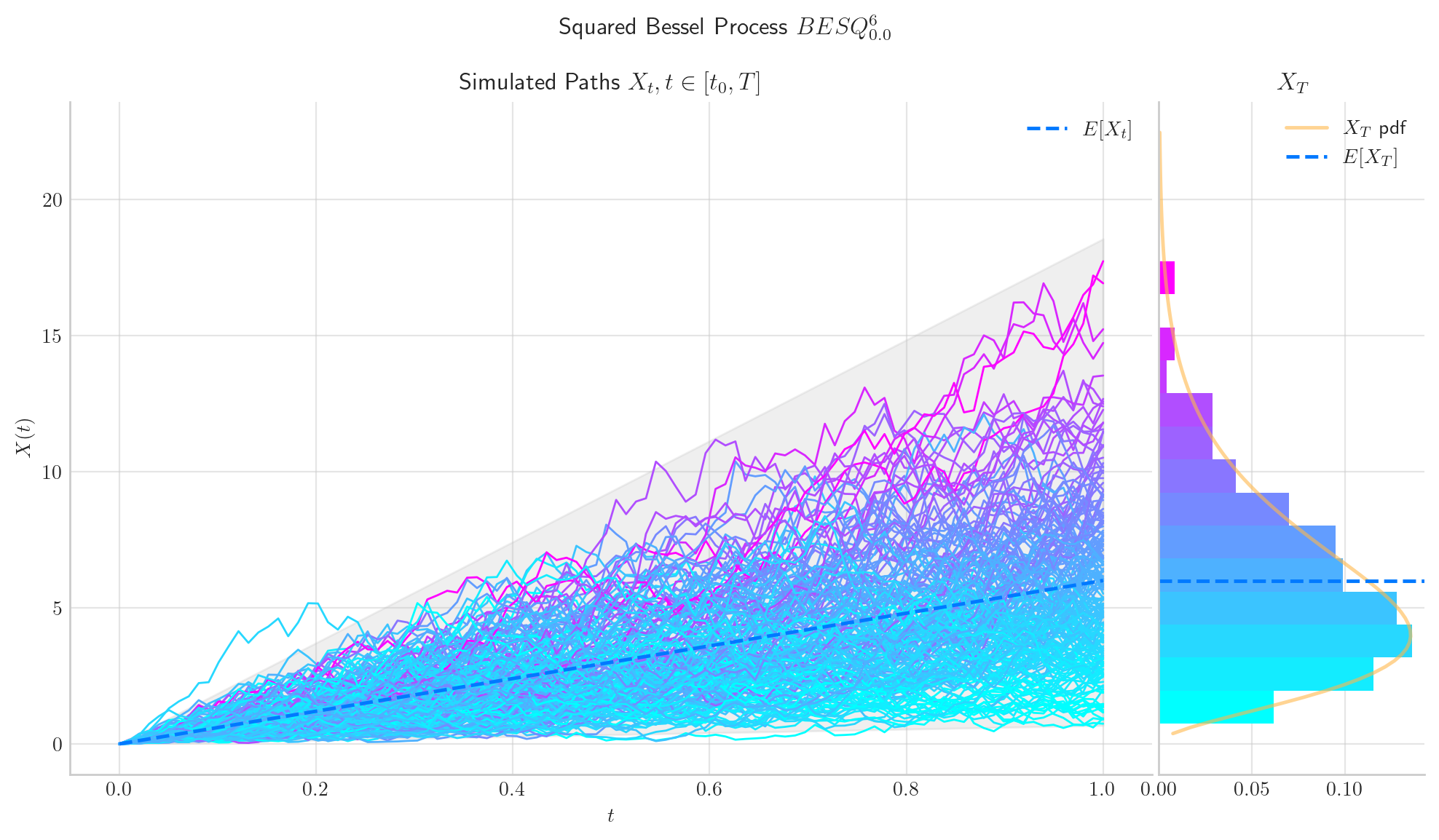
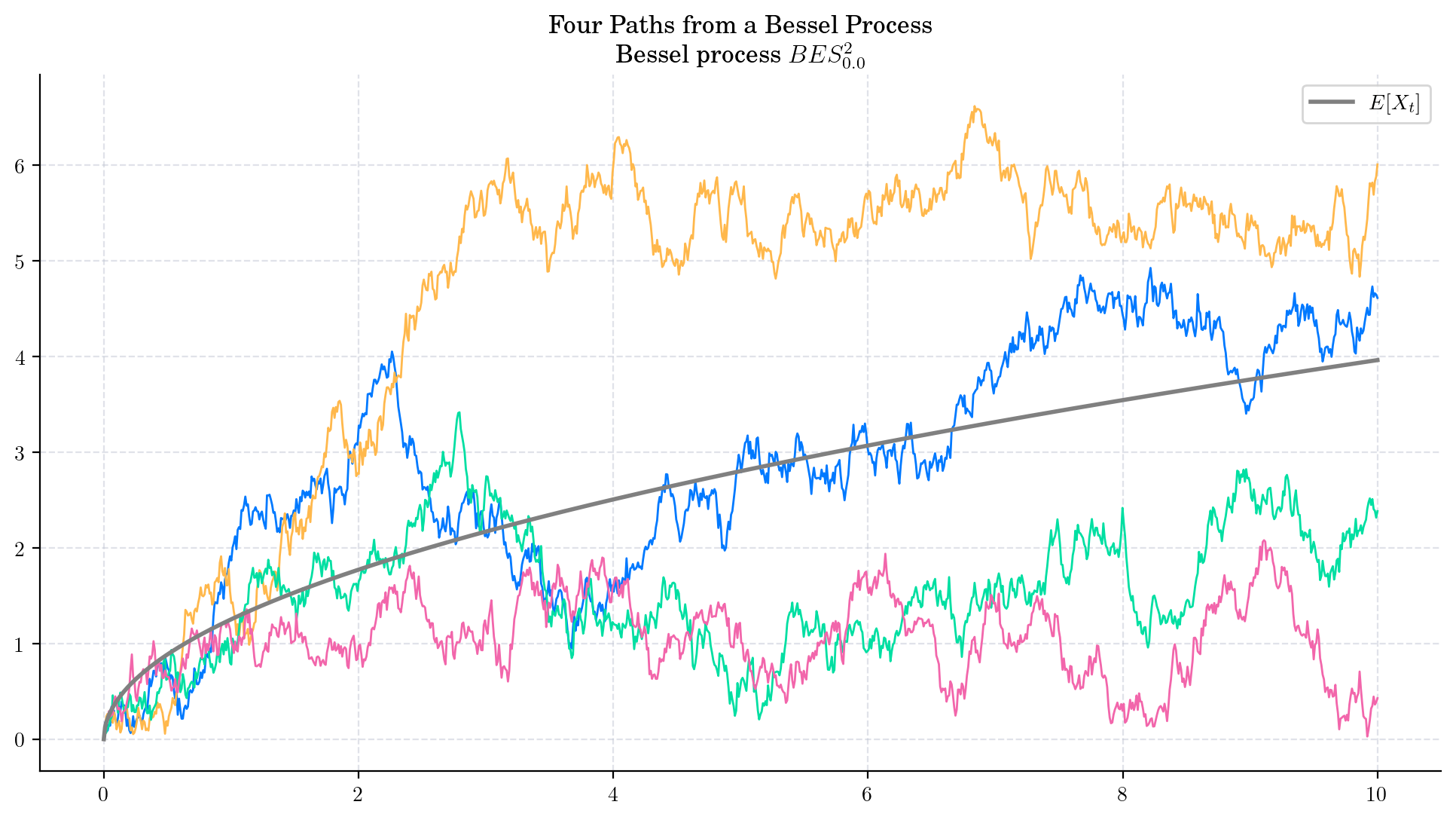
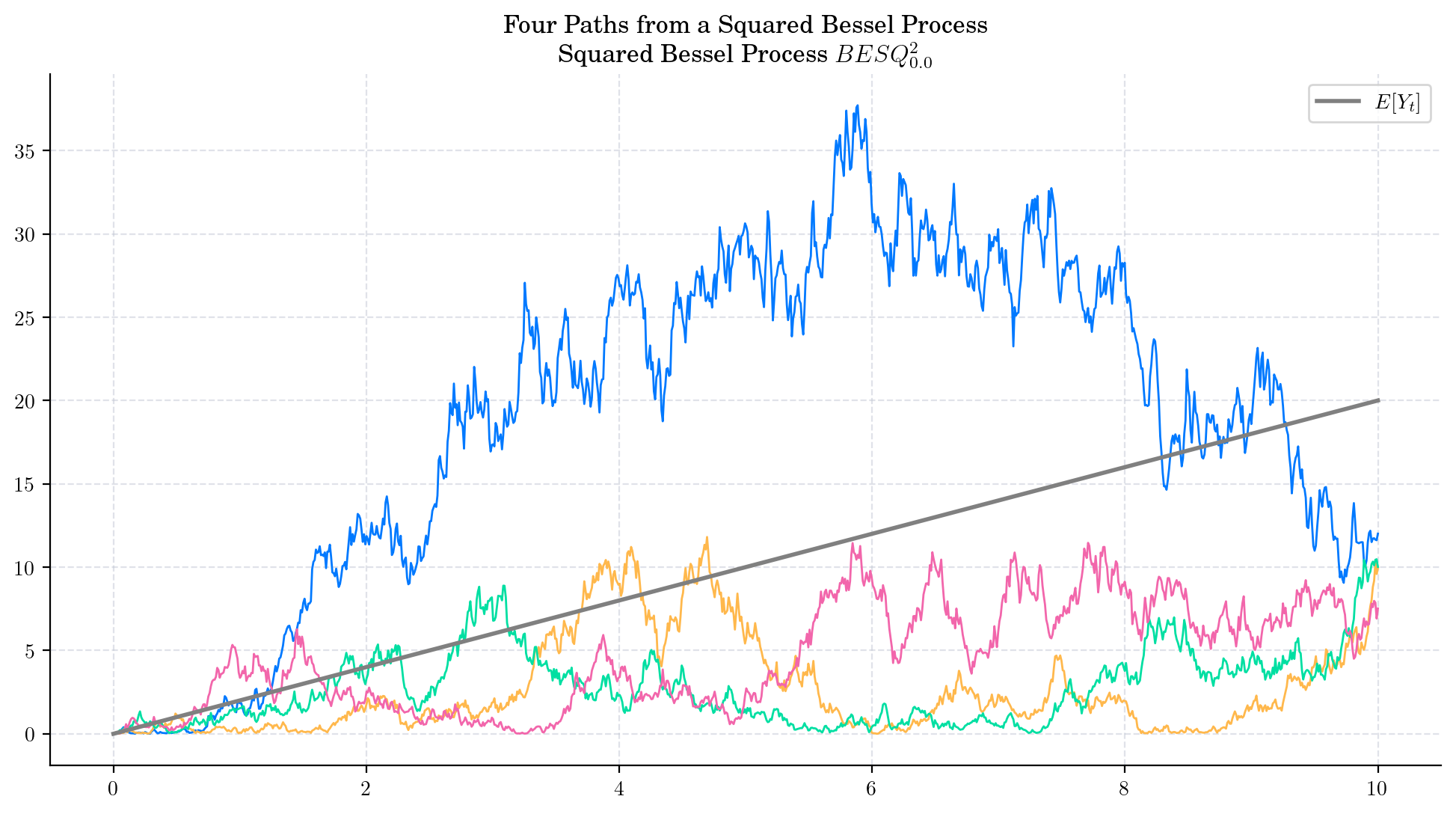
6.6. References and Further Reading#
“Mathematical Methods for Financial Markets” by Monique Jeanblanc, Marc Yor, Marc Chesney; Springer Science & Business Media, 13 Oct 2009
Notes on Bessel Processes by Gregory F. Lawler
A Survey and Some Generalizations of Bessel Processes by Anja Göing-Jaeschke and Marc Yor; Bernoulli Vol. 9, No. 2 (Apr., 2003), pp. 313-349 (37 pages)
Squared Bessel processes of positive and negative dimension embedded in Brownian local times by Jim Pitman, Matthias Winkel; Electron. Commun. Probab. 23: 1-13 (2018). DOI: 10.1214/18-ECP174
